schrodinger.livedesign.preprocessor module¶
Preprocess a molecule for ingestion into LiveDesign.
Preprocessing adjusts the input molecule to a standard form so that LiveDesign behaves in a consistent way. This adjusts both the depiction (e.g 2D layout) and the data associated with the molecule (properties, stereochemistry).
Customers often think of this step in terms of “deduplication”
- if I have two molecules, when should they be considered
“the same”? The answer is collected in
schrodinger.livedesign.registration.RegistrationOptions,
but much of it is implemented here.
Different customer sites have differing business logic, so most of this module is about supporting that diversity of different configurations.
We should encourage customers to stay as close to the default configuration as possible. The default is the most validated, and it generates the most consistent behavior for internal and external tools linked to LiveDesign.
Copyright Schrodinger, LLC. All rights reserved.
- class schrodinger.livedesign.preprocessor.ExplicitHydrogens¶
Bases:
enum.Enum- REMOVE_ALL = 1¶
Remove all hydrogens. Default; recommended for most users
- KEEP_WEDGED = 2¶
Keep only hydrogens that have wedge/dash bonds
- ADD_ALL = 3¶
Add hydrogens to all atoms
- AS_IS = 4¶
Preserve input hydrogens. Does not add or remove hydrogens
- ON_HETERO_AND_KEEP_WEDGED = 5¶
Add hydrogens to heteroatoms and keep hydrogens that have wedge/dash bonds
- class schrodinger.livedesign.preprocessor.GenerateCoordinates¶
Bases:
enum.Enum- NONE = 1¶
Use input coordinates
- FULL = 2¶
Generate totally new 2D coordinates/layout
- FULL_ALIGNED = 3¶
Generate totally new 2D coordinates/layout, but align with input coordinates. Default; recommended for most users
- class schrodinger.livedesign.preprocessor.ChiralFlag0Meaning¶
Bases:
enum.EnumSDF files have a “chiral flag” that can be 0 or 1. According to Biovia’s specification, chiral flag 0 means that the molecule refers to both itself and its enantiomer.
However, different companies have used the chiral flag to mean a variety of things (in part because the documentation of the file format postdates its actual use…)
- UNGROUPED_ARE_ABSOLUTE = 1¶
Ignore the Chiral Flag, treating all chiral centers as absolute. Default; recommended for most users
- UNGROUPED_ARE_RACEMIC = 2¶
Chiral flag 0 molecules are turned into AND (aka racemic) groups. This is the BIOVIA standard
- UNGROUPED_ARE_RELATIVE = 3¶
Chiral flag 0 molecules are turned into OR (aka unknown) groups
- class schrodinger.livedesign.preprocessor.PreprocessorOptions¶
Bases:
NamedTupleConfiguration options for the preprocessor. These options are typically stored in a json dict on a LiveDesign instance. Skipping a setting will use the default value for that setting.
- MAX_NUM_ATOMS: Optional[int]¶
Reject compounds with more than MAX_NUM_ATOMS atoms. Default: None (no limit)
- KEEP_ONLY_LARGEST_STRUCTURE: bool¶
Should additional discontiguous structures be removed? Default: True
- STRIP_SALTS: Optional[Tuple[str]]¶
Which salt molecules to avoid registering. Run after removal of small molecules. Default: None (no salts are removed)
- NEUTRALIZE: bool¶
Should the input be neutralized? Default: True
- TRANSFORMATIONS: Optional[Tuple[str]]¶
Standardize the representation of certain functional groups. Default transformations are:
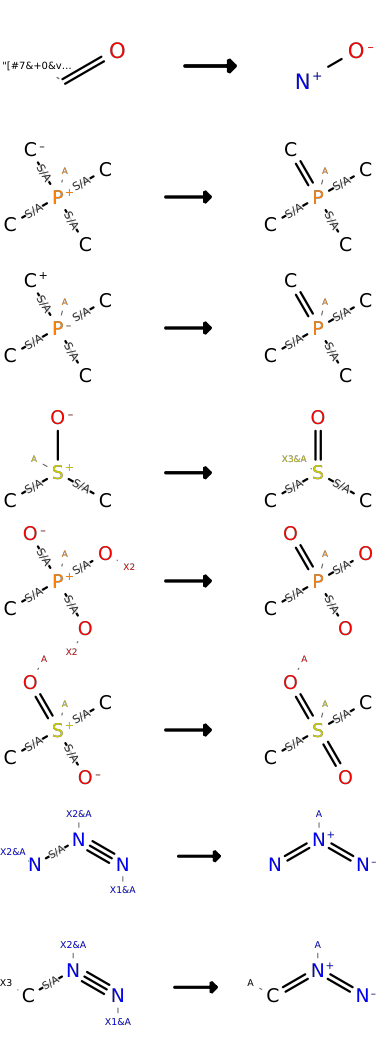
- CHOOSE_CANONICAL_TAUTOMER: bool¶
Canonicalize the tautomeric representation. Slow, but recommended if the TAUTOMER_INSENSITIVE_LAYERS hash scheme is chosen. Default: False
- RESOLVE_AMBIGUOUS_TAUTOMERS: bool¶
If the input includes aromatic or conjugated bonds, should we guess which tautomer was intended? If false, these compouunds will fail with a kekulization error, allowing the user to correct the input.
Default: False unless CHOOSE_CANONICAL_TAUTOMER=True
- CHIRAL_FLAG_0_MEANING: schrodinger.livedesign.preprocessor.ChiralFlag0Meaning¶
What does a chiral flag of 0 mean? Default: UNGROUPED_ARE_ABSOLUTE
- STRIP_AND_GROUPS_ON_SINGLE_ATOM: bool¶
If an AND group includes only one atom, remove the stereo annotation on that atom. Default: False
- PRESERVE_ENHANCED_STEREO_GROUP_IDS: bool¶
Preserve the group IDs of enhanced stereo groups. Default: False
- REMOVE_PROPERTIES: bool¶
Should properties be removed? Properties don’t affect deduplication, but the properties and s-group data from the first instance of a compound registered will be stored in LD. Default: False
- GENERATE_COORDINATES: schrodinger.livedesign.preprocessor.GenerateCoordinates¶
Should 2D coordinates be generated? Default: FULL_ALIGNED
- EXPLICIT_HYDROGENS: schrodinger.livedesign.preprocessor.ExplicitHydrogens¶
How should hydrogens be treated? Default: REMOVE_ALL
- CLEAR_INVALID_WEDGE_BONDS: bool¶
Should wedged bonds on achiral atoms be removed? Default: True
- WEDGE_TWO_BONDS_AROUND_CHIRAL_ATOMS: bool¶
Should we draw two wedged bonds on chiral atoms with 4 neighbors? Default: False
- HEAVY_HYDROGEN_DT: bool¶
Should the Deuterium and Tritium use symbols D and T? Default: False
- static fromConfig(config: dict)¶
Create a PreprocessorOptions object from a configuration dictionary in the format stored in LiveDesign.
Also rejects or updates deprecated options.
- Parameters
config – configuration from which to build options
- Raises
KeyError – if an unknown key is present
ValueError – if an unknown value is present
- toConfig() dict¶
Write a LiveDesign configuration dictionary from an options object
- schrodinger.livedesign.preprocessor.initialize_audit_log(verbose: bool)¶
Initialize global audit for logging purposes
- schrodinger.livedesign.preprocessor.remove_invalid_config_options(config: dict) Tuple[str]¶
- Parameters
config – configuration from which to build options, from which all invalid keys and values will be stripped
- Returns
tuple of errors encountered
- schrodinger.livedesign.preprocessor.audit_changes(func: Callable, mol: rdkit.Chem.rdchem.Mol, *args)¶
When the global audit_changes_log is initialized, compares mol CXSMILES before and after the given function call, capturing information when the CXSMILES has been changed.
- Parameters
func – transformation function
mol – molecule to apply transformation to
- schrodinger.livedesign.preprocessor.is_wildcard(atom)¶
Is atom a wildcard?
- schrodinger.livedesign.preprocessor.is_queryatom_exception(atom)¶
Normally we raise an exception if query atoms are in the molecule to be preprocessed, but we don’t want to do that if the atom is an attachment point
- Parameters
atom – the atom to check
- Returns
whether or not the atom is allowed in the preprocessor
- schrodinger.livedesign.preprocessor.coords_all_zero(conf)¶
Returns whether or not all atom positions in a conformer are zero
- schrodinger.livedesign.preprocessor.get_limited_sanitized_mol(mol: rdkit.Chem.rdchem.Mol) rdkit.Chem.rdchem.Mol¶
Sanitize the molecule in a limited way as to avoid changing the molecule or throwing when valence errors are present. Specifically we turn off: SANITIZE_PROPERTIES: which otherwise checks valences SANITIZE_CLEANUP: which can change the shape of molecule SANITIZE_CLEANUPCHIRALITY: which can remove chirality markers SANITIZE_FINDRADICALS: which checks valences of radicals
- schrodinger.livedesign.preprocessor.setup_mol(mol)¶
Setup on a molecule that is always done regardless of configuration.
- Parameters
mol – An unsanitized RDKit Mol
- Returns
A partially sanitized RDKit mol, ready for the standardizer.
- schrodinger.livedesign.preprocessor.check_kekulization(mol, options)¶
- schrodinger.livedesign.preprocessor.assign_zero_coords_chirality(mol)¶
Molecules with all-zero coordinates need to have the “chirality tags” primed from the atom parity properties. Once these are in place, we remove the conformer, and leave the mol in a state equivalent to one that came from a SMILES input.
- schrodinger.livedesign.preprocessor.correct_sgroup_coordinates(mol)¶
If coordinates are generated, make sure the FIELDDISP property in the Sgroups are using relative coordinates.
- schrodinger.livedesign.preprocessor.preprocess_molblock(molblock: str, config: Optional[dict] = None, preserved_data_sgroups: Optional[List[str]] = None) str¶
Standardizes an MDL molblock
- Parameters
molblock – input molblock
config – dict specifying preprocessor options
preserved_data_sgroups – list of sgroup names to preserve
- Returns
standardized molblock
- schrodinger.livedesign.preprocessor.preprocess(mol: rdkit.Chem.rdchem.Mol, options: Optional[schrodinger.livedesign.preprocessor.PreprocessorOptions] = None, preserved_data_sgroups: Optional[List[str]] = None) rdkit.Chem.rdchem.Mol¶
Standardizes an RDKit mol
- Parameters
mol – input mol
options – preprocessor options
preserved_data_sgroups – list of sgroup names to preserve
- Returns
standardized mol
- exception schrodinger.livedesign.preprocessor.BlindedCompoundError¶
Bases:
ValueError
- schrodinger.livedesign.preprocessor.assert_not_blinded(mol: rdkit.Chem.rdchem.Mol, max_num_atoms: Optional[int] = None)¶
Checks imcoming mol to confirm it has real atoms; if it doesn’t it may have been intentionally stripped by the caller. LiveDesign marks these structures as having been “blinded”, meaning a customer may have IP/legal restrictions, or there’s a delay in registering the structure despite having assay data available. Currently, LiveDesign handles these structures by keeping a row in the LiveReport, but without an associated SDF or image. This is different from other registration errors, which are simply archived.
- Parameters
mol – RDKit Mol to consider
max_mol_wt – maximum allowed molecular weight
- schrodinger.livedesign.preprocessor.assert_not_query(mol: rdkit.Chem.rdchem.Mol)¶
Checks incoming mol to confirm there are no query features present on atoms or bonds, which would otherwise make it not compatible with registration.
- Parameters
mol – RDKit Mol to consider
- schrodinger.livedesign.preprocessor.get_atoms_mapping(mol_atoms)¶
- schrodinger.livedesign.preprocessor.get_bonds_mapping(mol, original_bond_mapping, atom_idx_mapping)¶
- schrodinger.livedesign.preprocessor.tag_mol_indexes(mol)¶
Tag atoms with the initial indexes on the mol and create a bond mapping. Bonds are less stable than atoms, so we create an external mapping to the atoms they bind
- schrodinger.livedesign.preprocessor.check_attachment_points_changed(sg, atom_idx_mapping)¶
- schrodinger.livedesign.preprocessor.check_cstate_changed(sg, bond_idx_mapping)¶
- schrodinger.livedesign.preprocessor.update_sgroup_indexes(sg, sg_atoms, sg_parent_atoms, sg_bonds, atom_idx_mapping, bond_idx_mapping)¶
- schrodinger.livedesign.preprocessor.update_mol_groups(mol, stereo_groups, substance_groups, original_bond_mapping)¶
Update atoms and bonds in stereo and substance groups, dropping any atoms/groups that are no longer valid for the current state of the mol.
- schrodinger.livedesign.preprocessor.update_sgroups(mol, substance_groups, atom_idx_mapping, bond_idx_mapping)¶
Update SGroups to reflect the transformations done on mol, updating with new atom and bond indexes, as well as atoms that might have been added or removed.
- schrodinger.livedesign.preprocessor.update_stereo_groups(mol, stereo_groups, atom_idx_mapping)¶
- schrodinger.livedesign.preprocessor.add_explicit_hydrogens(mol, only_on_hetero=False)¶
- schrodinger.livedesign.preprocessor.remove_explicit_hydrogens(mol, sgroups, keep_wedged=False, keep_hetero=False)¶
- schrodinger.livedesign.preprocessor.convert_to_molblock(mol, options=None)¶
Convert processed mol into a molblock and make necessary updates.
- schrodinger.livedesign.preprocessor.convert_heavy_hydrogens(molblock)¶
NOTE that this operates on a molblock, not a molecule
The RDKit does not currently (v2020.03) support writing D or T to mol blocks, so we need to post-process the text. Fortunately it’s an easy regex in v3000 mol blocks. This does not work with V2000 mol blocks, so we throw a ValueError there. This doesn’t seem like a big deal since V2000 support is primarly being kept around for debugging purposes. If we need to eventually support V2000+HEAVY_HYDROGEN_DT, some not-completely-trivial code will need to be written.
- schrodinger.livedesign.preprocessor.neutralize(mol, checkForProblematicHs=False)¶
- schrodinger.livedesign.preprocessor.unicode_to_str(unicode_str)¶
Takes a unicode object and converts it to a str (utf-8). If the arg is already a str, returns unicode_str (i.e. if run with python 3). Needed to support python 2/3 with unicode_literals.
py2: type<unicode> -> type<str utf-8> py3: type<str utf-8> (no unicode type exists)
- Parameters
unicode_str (unicode (py2) or str (py3)) – the unicode that potentially needs converting (i.e. if run with python 2)
- Returns
str
- schrodinger.livedesign.preprocessor.transform(mol, transformation)¶
apply the transformation to the molecule repeatedly until it no longer applies.
the maxTransformations argument is just there to prevent us from ending up in an infinite loop due to a bogus transformation
Please note that running transformations may alter the stereochemistry of mol, so a stereo recalculation from coordinates might be required.
- schrodinger.livedesign.preprocessor.in_xy_plane(mol)¶
- schrodinger.livedesign.preprocessor.generate_coordinates(mol, align=False)¶
- schrodinger.livedesign.preprocessor.is_polymer(s_group)¶
- schrodinger.livedesign.preprocessor.clean_up_polymer_brackets(mol, revert_to_mol=None, keep_existing_brackets=False)¶
Add polymer brackets back to mol.
- Parameters
mol – RDKit mol to add polymer brackets to
revert_to_mol – RDKit mol to revert to if polymer brackets cannot be added correctly to provided mol. This will occur when brackets cross more than one bond.
keep_existing_brackets – whether to recalculate the positions of brackets that are already present
- Returns
RDKit mol with polymer brackets
- schrodinger.livedesign.preprocessor.copy_lewis_structure_and_hydrogens(st, mol)¶
Applies bond orders and charges from st to mol. Updates #implicitH to match
Assumes st includes all hydrogens. Adds implicit and explicit hydrogens to the mol, but does not add any graph hydrogens. May remove graph hydrogens.
- schrodinger.livedesign.preprocessor.generate_canonical_tautomer(mol)¶
- schrodinger.livedesign.preprocessor.clear_wedge_bonds_from_achiral_centers(mol)¶
- schrodinger.livedesign.preprocessor.calculate_enhanced_stereo(mol, enh_stereo_default_grouping, initial_chiral_flag)¶
- schrodinger.livedesign.preprocessor.strip_stereo_and(input_mol)¶
Removes any Stereo AND groups with only one center and flattens the bonds around it
- Parameters
input_mol – The original molecule to consider
- Returns
post-processed molecule, if the input molecule was modified
- schrodinger.livedesign.preprocessor.frag_is_smaller(atoms, largest_atoms, weight, largest_weight, smiles, largest_smiles)¶
A fragment is considered larger if its atoms/weight are larger, the length of the smiles string is larger, or the smiles string is lexicographically smaller if they are equal length. ie, ‘AAA’ is larger than ‘AAB’.. hence the final smiles > largest_smiles check here to reject
- schrodinger.livedesign.preprocessor.connect_variable_attachment_points(mol)¶
forms zero-order bonds between one of the atoms of a bond with an ATTACH property to the “main” molecule in order to have the molecule+variable attachment point treated as a single fragment
- returns a 2-tuple with:
the modified molecule
whether or not the molecule was modified
- schrodinger.livedesign.preprocessor.remove_fragments(mol, substance_groups)¶
Fragments are not removed if the molecule contains any SGroups which are associated with polymers
- Use the following criteria to remove unwanted fragments from mol:
keep only the fragment which has the most number of atoms
break ties by keeping only fragments with the greatest molecular weight
break ties with the longest smiles string
break additional ties by keeping the fragment with the earliest alpha sorted SMILES string
If two or more identical fragments remain after 1-4, we will throw a fatal error.
- schrodinger.livedesign.preprocessor.remove_properties(mol)¶
- schrodinger.livedesign.preprocessor.strip_salts(mol, salt_list)¶
- schrodinger.livedesign.preprocessor.apply_transformations(mol, transformations)¶
Apply the given list of transformations, and recalculate stereo if at least one transformation applies.
- schrodinger.livedesign.preprocessor.add_chiral_hs(mol)¶
- schrodinger.livedesign.preprocessor.wedge_clean(mol, wedge_2_bonds_if_possible)¶
- schrodinger.livedesign.preprocessor.remove_wiggly_bonds_around_double_bonds(mol)¶